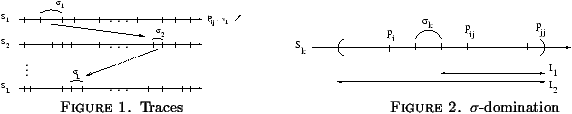Nearest-Neighbour Search in High Dimension and Molecular Clustering
Frédéric Cazals
Algorithms project, INRIA Rocquencourt
Algorithms Seminar
June 30, 1997
[summary by Frédéric Cazals]
A properly typeset version of this document is available in
postscript and in
pdf.
If some fonts do not look right on your screen, this might be
fixed by configuring your browser (see the documentation here).
1 Introduction and prerequisites
1.1 Problem statement
Given a set of points P={p1,...,pn} Ì Rd, the
nearest-neighbour (NN) and k nearest neighbours (k-NN) problems
can be stated as follows: pre-process P in order to return as fast
as possible the nearest or k nearest neighbour(s) of an arbitrary
point q according to any Euclidian metric d(p,q) = (åi=1d
(pi - qi)2)1/2. A weakened version of the NN problem consists
in returning a point p' which e-approximates the NN p of
q in the sense d(p',q) d(p,q) £ 1+e for any e >
0. If one denotes pi1,...,pin the points of P sorted
by increasing distance to q, an equivalent formulation for the
k-NN problem consists in returning a subset S={s1,...,sk}
with d(q,sj) £ (1+e)d(q,pij) for j=1,...,k.
The naive algorithm to compute the NN of a point q consists in
checking all the points of P and returning the closest, which has
complexity O(dn). On the other hand, the most sophisticated
algorithms known until recently had complexities in O(exp(d) log n)
with exp(d) a function growing at least as quickly as 2d---see
e.g., [1]. So that whenever d ³ log n nothing better
than the brute force method was known!
Kleinberg's break-through [4] has been to get around the
exponential difficulty by an heavy use of random sampling aiming at
``comparing'' the points of P through their projections on random
lines passing through the origin rather than decomposing the
d-dimensional space containing them. The first result is an
algorithm returning an approximation of the k-NN in a deterministic
way but with an exponential time/space pre-processing. The second
algorithm returns an approximation of the NN in a randomised way but
with a polynomial pre-processing only. This talk presents these two
algorithms and discusses their potential use to a clustering problem
arising in chemistry---see section 4.
1.2 Prerequisites
1.2.1 A geometric lemma
The core idea of Kleinberg's method lies in the following property:
Lemma 1
Let x and y be two vectors of Rd such that ||y||/||x||
³ 1+g with g £ 1/2. Then, if v is a random
vector on the unit sphere Sd-1 we have
Pr[|x · v| ³ |y · v|] £ 1/2 - g/3.
Intuitively, short vectors ``should not defeat too often'' longer ones
when comparing their projection on a random line determined by a
vector on Sd-1. In order to compare two vectors from their
projections, the key point is therefore to use a large enough set of
lines to capture the probabilistic property contained in the above
theorem.
1.2.2 Empirical measures and Vapnik-Chervonenkis bounds
Let (W, F,µ) be a probability space, S Ì
F a set of events, and X1,X2,...,Xn n random
variables following the law µ. If one calls the empirical measure
of an event s the fraction of Xi's falling into s, the quantity
measures the maximum difference over the class S between the
empirical measure and the probability. It is a random variable and
Vapnik-Chervonenkis's contribution [5] has been to
elucidate the conditions under which it converges in probability to
zero, that is the conditions under which
limn ® ¥ Pr[DSn > e] = 0
for any e.
To sketch this contribution, let a range-space be a couple ( P, R)=
((W, F,µ), R Ì F
).
We shall say that a set A of finite cardilality is shattered by
R if " a Î 2A $ rÎ R
such that a=r Ç A. The dimension of Vapnik-Chervonenkis of
( P, R) is the cardinality of the biggest A Ì W
shattered by R. Definition 1
A g-sample for ( P, R) is a finite set A Ì W
such that
|µ(r)-| r È A |/| A || £ g,
" rÎ R.
Theorem 1 [[5]]
For a range space of dimension k, a random sample of size
is a g-sample with a probability at least 1-d.
1.2.3 Exceptional and r-distinguishing sets
As pointed out above, we are interested in comparing points with
respect to their projections on vectors from Sd-1. For two
vectors x and y with ||y|| / ||x|| ³ 1+g
we call their exceptional set
| W |
|
= { v Î Sd-1 such that
| x · v| ³ | y · v |
}.
|
| " W |
|
, µ(W |
|
)<r
Þ
| V Ç W |
|
| / | V | < 1/2.
|
1.2.4 Hyper-planes arrangements
An arrangement of n hyper-planes in Rd is said to be in general
position if any d hyper-planes have a unique point in common, and
any d+1 hyper-planes do not share a point. Given a hyper-plane h
and a point p, p is either above, on, or below h, which is
called its position. A face on an arrangement is the set of points
having the same position with respect to all the hyper-planes. The
dimension of a face is its affine dimension. It is known from [2] that
Theorem 2
The number of d-faces of an arrangement of n hyper-planes in
general position is fd(n)=åi=0d ().
1.2.5 Digraphs
A complete digraph G on n vertices 1,2,...,n is a directed
graph which contains for any pair of vertices {i,j} either the
edge (i,j) or (i,j). An apex of G is a vertex with a directed
path of length at most two to any vertex. At least, an apex ordering
of G is an ordering i1,...,in of its vertices such that
ik is an apex for the sub-digraph G[ik,ik+1,...,in]. The
following is straightforward:
Theorem 3
Every n-node complete digraph has an apex ordering computable in
O(n2).
1.3 First results
The following theorems can be proved:
Lemma 2
Let µ be the uniform measure on Sd-1. The dimension of the
range-space
| ((Sd-1, F |
, µ), { W |
|
|µ(W |
|
) £ r }) |
Lemma 3
There exists c0 such that a random sample of Sd-1 of size
f(d,g) is a g/2-sample for the range-space
((Sd-1, F, µ), { Wx,y|µ(Wx,y) £ r })
with
f(d,g)=
c0/g2
(d' log d'/g2 + log 1/d)=
q(d log2 d).
Corollary 1
A set V of f(g,d) vectors from Sd-1
is (1/2-g)-distinguishing with a probability
at least 1-d.
2 First algorithm
2.1 Construction of the data structure
This algorithm returns an approximation of the k-NN of a point q.
To build the data structure from which it does so, we first draw
uniformly at random a set V of L = f(e/3,d) =
q(d log2 d) vectors from Sd-1. Then for each vector vl
Î V, the following is done: 1. compute vl · pij with
pij=(pi+pj)/2, 1£ i,j£ n; 2. sort the
pij according to the values of vl · pij and denote
Sl the list obtained. The list of lists S1,...,SL is denoted
S.
For each such list, a pair of consecutive entries is called a
primitive interval, and a sequence of primitive intervals is called a
trace---see Figure 1. The maximum number of
traces is upper-bounded by (n2)L = nO(d log2 d). But a trace
is realizable if
| $ q Î Rd, "
k=1,...,L,
vk · |
p |
|
< vk · q
< vk · |
p |
|
.
|
Definition 2
For a realizable trace s=s1 ··· sk ···
sL, pi is said to s-dominate pj in Sk if
pij lies in the interval (sk,...,pjj).
For each realizable trace s, the construction is as follows:
(1.) build a complete digraph Gs on {1,2,...,n} with
the edge (i,j) if pi s-dominates pj in half of the
lists of S; (2.) build an apex ordering (s,Gs) of
the nodes of Gs.
The idea behind the domination definition is depicted on Figure
2: if pij falls in the desired interval
denoted I1, then pi Î I2 and we have
|vk · (pi-q)| < |vk · (pj-q)|.
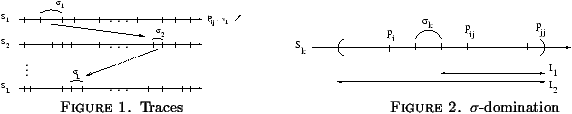
2.2 Algorithm
To process a query associated to a point q: 1. compute
s(q)=s1(q) ··· sL(q) with sk(q) the
primitive interval from Sl containing vl · q; 2. retrieve
the apex ordering associated to s(q) and return the k first
entries.
This algorithm therefore returns an e-approximation of the k
nearest neighbours of a point q in a deterministic fashion, so that
the answer is guaranteed to be correct if the random sample V is
actually (1/2-e/3)-distinguishing---see §1.2.3
and Corollary 1.
Roughly speaking, the correctness of the algorithm comes from the fact
that if a non-desired point p1 has been returned instead of a
desired point p2, then p1 dominated p2 in more than half of
the lists of S, and the sample V was not distinguishing
enough with respect to the exceptional set Wq-p1,q-p2.
The pre-processing requires O (Ln2 (n log d)(2d) )
time and O (n (n log d)(2d) ) space. The cost of a
query is O(k+L (d+log n))=O(k + (d log2 d) (d + log n)).
3 Second algorithm
As opposed to the first algorithm, the second one does not try to
compute a partition of the requests' space and proceeds in two
steps. The first one consists in drawing a random sample from P of
the appropriate size and returning the closest point to q. The
second one compares iteratively q to pairs of points of P in a
tournament way depicted on Figure 3. Assuming that
n=2m, the overview of this second stage is the following:
-
a random sample V of Q(d log2 n (log2 d + log d
log log n)) vectors is drawn uniformly on Sd-1, and a
multi-set Gv of V is assigned to each internal node v of
the binary tree whose leaves are a permutation of the points of P.
For a query point q, two points pi and pj of P, and a
multi-set Gv, pi is said to dominate pj if
|vk · (pi-q)| < |vk · (pj-q)|
holds for a majority of vectors vk in Gv. Otherwise pj
dominates pi;
- each internal node v of the tree is assigned its dominating
child for the multi-set Gv.
The point eventually returned is the best candidate from the two
points returned by the two steps. It can be shown that an
e-approximation is returned with a probability greater than
1-d in O(n + d log3 n) time with a space requirement of
n| V |.
Figure 1: Tournament tree
Figure 2: Molecule and fragments

4 Application to molecular clustering
Suppose we are given a set of d molecular fragments, say
F={f1,...,fd} and a set M={m1,...,mNm} of Nm
molecules, each described by a set of fragments of F---see
Figure 4. We shall represent a molecule mi by a sequence
of d boolean values mi=bi, 1 bi, 2 ··· bi,d with
bi, j=1 if mi contains fj and bi, j=0 otherwise. This
formulation fails to report multiple occurrences of a given fragment
in a molecule, but it has the nice property that a molecule is
represented by a point on the hyper-cube Hd={0,1}d. Given two
molecules, we call their similarity the number of common
fragments that is the quantity sim(mi,mj) =
åk=1d bi, k· bj, k.
Given the set M we are interested in the following problem: find a
partition of M into subsets of neighbours or clusters. To do so,
one way to proceed see [3] consists in first building a
Minimum Spanning Tree on the input data set, second removing those
``too long'' edges from the MST, and third computing the connected
components we are left with. The key point lies in the MST
computation, and it is shown in [6] that
Theorem 4
A MST on n points in dimension d can be found in
O(2dn2-1/2d+1 (log n)1-1/2d+1) time.
Unfortunately for our concern where d can range from 500 to 2000,
the 2d constant is prohibitive. Kleinberg's algorithms customised
to the hyper-cube setting could make Yao's algorithm interesting in
practice.
References
- [1]
-
Arya (S.), Mount (D. M.), Netanyahu (N. S.), Silverman (R.), and Wu (A. Y.). --
An optimal algorithm for approximate nearest neighbour searching. In
Proceedings of the Fifth Annual ACM-SIAM Symposium on Discrete
Algorithms. pp. 573--582. --
New York, 1994.
- [2]
-
Edelsbrunner (Herbert). --
Algorithms in combinatorial geometry. --
Springer-Verlag, Berlin, 1987, EATCS Monographs
on Theoretical Computer Science, vol. 10, xvi+423p.
- [3]
-
Jain (Anil K.) and Dubes (Richard C.). --
Algorithms for clustering data. --
Prentice-Hall Inc., Englewood Cliffs, NJ, 1988, Prentice-Hall Advanced Reference Series, xiv+320p.
- [4]
-
Kleinberg (J.). --
Two algorithms for nearest-neighbour search in high dimension. In
ACM STOC. --
El Paso, Texas, USA, 1997.
- [5]
-
Vapnik (N.) and Chervonenkis (A.). --
On the uniform convergence of relative frequencies of events to their
probabilities. Theory of Probability and its Applications, vol. 16,
n°2, 1971, pp. 264--280.
- [6]
-
Yao (Andrew Chi Chih). --
On constructing minimum spanning trees in k-dimensional spaces and
related problems. SIAM Journal on Computing, vol. 11, n°4,
1982, pp. 721--736.
This document was translated from LATEX by
HEVEA.